Crystal Growth Simulation – Part 1
Ever since I read chapter 2 of Fabrication Engineering at the Micro- and Nanoscale, I have been interested in simulating crystal growth, primarily because of the multi-discplinary nature of this problem: finite element modeling software, modeling using high performance computing, the physics of the problem (governing laws and equations), and visualization techniques. I had run across Elmer when looking up crystal growth examples a while back. About two weeks ago, I was watching this Elmer demo in which they used gmsh to show the geometry for the simulation.
I wanted to learn more about Gmsh and how it is used, so I took a detour into this Gmsh introductory video. It was an excellent tour of Gmsh and its features.
Gmsh for Crystal Growth
At this point, I wanted to see how Gmsh could be used for crystal growth simulation so I searched for “crystal growth gmsh“. I was very pleased to find this paper: Development and validation of a thermal simulation for the Czochralski crystal growth process using model experiments – ScienceDirect. It appeared to have a corresponding set of slides and a talk as well. It especially piqued my interest that there is a script to set up a mesh for a Czochralski furnace.
Running the Code
Thankfully, the code in this paper is open source. I already had python installed so the first thing I tried was executing run.py in Git bash
$ git clone https://github.com/nemocrys/test-cz-induction
$ cd test-cz-induction
$ python run.py
Traceback (most recent call last):
File "D:\dev\repos\fem\research\test-cz-induction\run.py", line 5, in <module>
import opencgs.control as ctrl
ModuleNotFoundError: No module named 'opencgs'Without using my brain, I decided to install pyelmer since I knew it was a dependency (from the talk).
$ pip install pyelmer
Collecting pyelmer
Downloading pyelmer-1.0.0-py3-none-any.whl (27 kB)
Collecting gmsh
Downloading gmsh-4.10.5-py2.py3-none-win_amd64.whl (38.4 MB)
|████████████████████████████████| 38.4 MB 285 kB/s
Collecting pyyaml
Downloading PyYAML-6.0-cp310-cp310-win_amd64.whl (151 kB)
|████████████████████████████████| 151 kB ...
Collecting matplotlib
Downloading matplotlib-3.5.2-cp310-cp310-win_amd64.whl (7.2 MB)
|████████████████████████████████| 7.2 MB 6.4 MB/s
Collecting pillow>=6.2.0
Downloading Pillow-9.2.0-cp310-cp310-win_amd64.whl (3.3 MB)
|████████████████████████████████| 3.3 MB ...
Collecting cycler>=0.10
Downloading cycler-0.11.0-py3-none-any.whl (6.4 kB)
Collecting numpy>=1.17
Downloading numpy-1.23.1-cp310-cp310-win_amd64.whl (14.6 MB)
|████████████████████████████████| 14.6 MB 6.4 MB/s
Collecting packaging>=20.0
Downloading packaging-21.3-py3-none-any.whl (40 kB)
|████████████████████████████████| 40 kB 2.5 MB/s
Collecting python-dateutil>=2.7
Downloading python_dateutil-2.8.2-py2.py3-none-any.whl (247 kB)
|████████████████████████████████| 247 kB ...
Collecting kiwisolver>=1.0.1
Downloading kiwisolver-1.4.4-cp310-cp310-win_amd64.whl (55 kB)
|████████████████████████████████| 55 kB 1.6 MB/s
Collecting pyparsing>=2.2.1
Downloading pyparsing-3.0.9-py3-none-any.whl (98 kB)
|████████████████████████████████| 98 kB 6.8 MB/s
Collecting fonttools>=4.22.0
Downloading fonttools-4.34.4-py3-none-any.whl (944 kB)
|████████████████████████████████| 944 kB 6.4 MB/s
Collecting six>=1.5
Downloading six-1.16.0-py2.py3-none-any.whl (11 kB)
Installing collected packages: six, pyparsing, python-dateutil, pillow, packaging, numpy, kiwisolver, fonttools, cycler, pyyaml, matplotlib, gmsh, pyelmer
WARNING: Failed to write executable - trying to use .deleteme logic
ERROR: Could not install packages due to an OSError: [WinError 2] The system cannot find the file specified: 'C:\\Python310\\Scripts\\f2py.exe' -> 'C:\\Python310\\Scripts\\f2py.exe.deleteme'
WARNING: You are using pip version 21.2.4; however, version 22.2 is available.
You should consider upgrading via the 'C:\Python310\python.exe -m pip install --upgrade pip' command.Looks like I need to run this as an administrator.
C:\dev>pip install pyelmer
Collecting pyelmer
Using cached pyelmer-1.0.0-py3-none-any.whl (27 kB)
Collecting pyyaml
Using cached PyYAML-6.0-cp310-cp310-win_amd64.whl (151 kB)
Collecting gmsh
Using cached gmsh-4.10.5-py2.py3-none-win_amd64.whl (38.4 MB)
Collecting matplotlib
Using cached matplotlib-3.5.2-cp310-cp310-win_amd64.whl (7.2 MB)
Requirement already satisfied: python-dateutil>=2.7 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (2.8.2)
Requirement already satisfied: packaging>=20.0 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (21.3)
Requirement already satisfied: fonttools>=4.22.0 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (4.34.4)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (1.4.4)
Requirement already satisfied: pillow>=6.2.0 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (9.2.0)
Requirement already satisfied: pyparsing>=2.2.1 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (3.0.9)
Requirement already satisfied: numpy>=1.17 in c:\python310\lib\site-packages (from matplotlib->pyelmer) (1.23.1)
Collecting cycler>=0.10
Using cached cycler-0.11.0-py3-none-any.whl (6.4 kB)
Requirement already satisfied: six>=1.5 in c:\python310\lib\site-packages (from python-dateutil>=2.7->matplotlib->pyelmer) (1.16.0)
Installing collected packages: cycler, pyyaml, matplotlib, gmsh, pyelmer
Successfully installed cycler-0.11.0 gmsh-4.10.5 matplotlib-3.5.2 pyelmer-1.0.0 pyyaml-6.0
WARNING: You are using pip version 21.2.4; however, version 22.2 is available.
You should consider upgrading via the 'C:\Python310\python.exe -m pip install --upgrade pip' command.Installing pyelmer is not sufficient to avoid the “No module named ‘opencgs’” error. The solution is to clone the opencgs repo and use pip to install (as administrator) from that repo.
git clone https://github.com/nemocrys/opencgs
cd opencgsD:\...\research\opencgs>pip install -e .
Obtaining file:///D:/dev/repos/fem/research/opencgs
Collecting meshio
Using cached meshio-5.3.4-py3-none-any.whl (167 kB)
Collecting pandas
Using cached pandas-1.4.3-cp310-cp310-win_amd64.whl (10.5 MB)
Requirement already satisfied: pyyaml in c:\python310\lib\site-packages (from opencgs==0.3.1) (6.0)
Requirement already satisfied: pyelmer in c:\python310\lib\site-packages (from opencgs==0.3.1) (1.0.0)
Collecting rich
Using cached rich-12.5.1-py3-none-any.whl (235 kB)
Requirement already satisfied: numpy in c:\python310\lib\site-packages (from meshio->opencgs==0.3.1) (1.23.1)
Requirement already satisfied: python-dateutil>=2.8.1 in c:\python310\lib\site-packages (from pandas->opencgs==0.3.1) (2.8.2)
Collecting pytz>=2020.1
Using cached pytz-2022.1-py2.py3-none-any.whl (503 kB)
Requirement already satisfied: six>=1.5 in c:\python310\lib\site-packages (from python-dateutil>=2.8.1->pandas->opencgs==0.3.1) (1.16.0)
Requirement already satisfied: matplotlib in c:\python310\lib\site-packages (from pyelmer->opencgs==0.3.1) (3.5.2)
Requirement already satisfied: gmsh in c:\python310\lib\site-packages (from pyelmer->opencgs==0.3.1) (4.10.5)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\python310\lib\site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (1.4.4)
Requirement already satisfied: pyparsing>=2.2.1 in c:\python310\lib\site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (3.0.9)
Requirement already satisfied: cycler>=0.10 in c:\python310\lib\site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (0.11.0)
Requirement already satisfied: pillow>=6.2.0 in c:\python310\lib\site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (9.2.0)
Requirement already satisfied: fonttools>=4.22.0 in c:\python310\lib\site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (4.34.4)
Requirement already satisfied: packaging>=20.0 in c:\python310\lib\site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (21.3)
Collecting commonmark<0.10.0,>=0.9.0
Using cached commonmark-0.9.1-py2.py3-none-any.whl (51 kB)
Requirement already satisfied: pygments<3.0.0,>=2.6.0 in c:\python310\lib\site-packages (from rich->meshio->opencgs==0.3.1) (2.12.0)
Installing collected packages: commonmark, rich, pytz, pandas, meshio, opencgs
Running setup.py develop for opencgs
Successfully installed commonmark-0.9.1 meshio-5.3.4 opencgs-0.3.1 pandas-1.4.3 pytz-2022.1 rich-12.5.1
WARNING: You are using pip version 21.2.4; however, version 22.2 is available.
You should consider upgrading via the 'C:\Python310\python.exe -m pip install --upgrade pip' command.Trying to run gives an error about a missing module named objectgmsh. My assumption was the installing pyelmer was supposed to bring in all such modules.
D:\dev\repos\fem\research\test-cz-induction>python run.py
Traceback (most recent call last):
File "D:\dev\repos\fem\research\test-cz-induction\run.py", line 5, in <module>
import opencgs.control as ctrl
File "d:\dev\repos\fem\research\opencgs\opencgs\__init__.py", line 3, in <module>
import opencgs.geo
File "d:\dev\repos\fem\research\opencgs\opencgs\geo.py", line 1, in <module>
from objectgmsh import *
ModuleNotFoundError: No module named 'objectgmsh'I notice while reviewing the paper yet again that it mentions the setup being in a Dockerfile. Sure enough, objectgmsh is a separate package that needs to be installed using pip. I upgrade pip for good measure since those warnings are getting annoying.
\test-cz-induction>pip install objectgmsh
Collecting objectgmsh
Downloading objectgmsh-0.9-py3-none-any.whl (25 kB)
Requirement already satisfied: gmsh in c:\python310\lib\site-packages (from objectgmsh) (4.10.5)
Requirement already satisfied: numpy in c:\python310\lib\site-packages (from objectgmsh) (1.23.1)
Installing collected packages: objectgmsh
Successfully installed objectgmsh-0.9I realize I should probably be running setup.py instead of run.py. Here’s the output now!
\test-cz-induction>python setup.py
crucible
melt
crystal
inductor
seed
insulation
crucible_adapter
axis_bt
vessel
axis_top
crucible 0.001999999999999999
melt 0.003
crystal 0.0004
inductor 0.001
seed 0.00017999999999999998
insulation 0.005
crucible_adapter 0.0039
axis_bt 0.0025
vessel 0.0025
axis_top 0.0025
filling 0.0263
Warning: Mesh size = 0 for bnd_melt. Ignoring this shape...
Warning: Mesh size = 0 for bnd_seed. Ignoring this shape...
Warning: Mesh size = 0 for bnd_crystal_side. Ignoring this shape...
Warning: Mesh size = 0 for bnd_crystal_top. Ignoring this shape...
Warning: Mesh size = 0 for bnd_axtop_side. Ignoring this shape...
Warning: Mesh size = 0 for bnd_axtop_bt. Ignoring this shape...
Warning: Mesh size = 0 for bnd_crucible_bt. Ignoring this shape...
Warning: Mesh size = 0 for bnd_crucible_outside. Ignoring this shape...
Warning: Mesh size = 0 for bnd_ins. Ignoring this shape...
Warning: Mesh size = 0 for bnd_adp. Ignoring this shape...
Warning: Mesh size = 0 for bnd_axbt. Ignoring this shape...
Warning: Mesh size = 0 for bnd_vessel_inside. Ignoring this shape...
Warning: Mesh size = 0 for bnd_vessel_outside. Ignoring this shape...
Warning: Mesh size = 0 for bnd_inductor_outside. Ignoring this shape...
Warning: Mesh size = 0 for bnd_inductor_inside. Ignoring this shape...
Warning: Mesh size = 0 for bnd_symmetry_axis. Ignoring this shape...
Warning: Mesh size = 0 for if_crucible_melt. Ignoring this shape...
Warning: Mesh size = 0 for if_melt_crystal. Ignoring this shape...
Warning: Mesh size = 0 for if_crystal_seed. Ignoring this shape...
Warning: Mesh size = 0 for if_seed_axtop. Ignoring this shape...
Warning: Mesh size = 0 for if_axtop_vessel. Ignoring this shape...
Warning: Mesh size = 0 for if_crucible_ins. Ignoring this shape...
Warning: Mesh size = 0 for if_ins_adp. Ignoring this shape...
Warning: Mesh size = 0 for if_adp_axbt. Ignoring this shape...
Warning: Mesh size = 0 for if_axbt_vessel. Ignoring this shape...
Info : Meshing 1D...
Info : [ 0%] Meshing curve 3 (Line)
Info : [ 10%] Meshing curve 4 (Line)
Info : [ 10%] Meshing curve 15 (Line)
Info : [ 10%] Meshing curve 16 (Line)
Info : [ 10%] Meshing curve 17 (Line)
Info : [ 10%] Meshing curve 18 (BSpline)
Info : [ 20%] Meshing curve 19 (Circle)
...
Info : [100%] Meshing surface 12 order 2
Info : [100%] Meshing surface 13 order 2
Info : [100%] Meshing surface 14 order 2
Info : Surface mesh: worst distortion = 0.526353 (0 elements in ]0, 0.2]); worst gamma = 0.252838
Info : Done meshing order 2 (Wall 0.990998s, CPU 0.75s)
-------------------------------------------------------
Version : 4.10.5
License : GNU General Public License
Build OS : Windows64-sdk
Build date : 20220701
Build host : gmsh.info
Build options : 64Bit ALGLIB[contrib] ANN[contrib] Bamg Blas[petsc] Blossom Cgns DIntegration DomHex Eigen[contrib] Fltk Gmm[contrib] Hxt Jpeg Kbipack Lapack[petsc] MathEx[contrib] Med Mesh Metis[contrib] Mmg Mpeg Netgen NoSocklenT ONELAB ONELABMetamodel OpenCASCADE OpenCASCADE-CAF OpenGL OpenMP OptHom PETSc Parser Plugins Png Post QuadMeshingTools QuadTri Solver TetGen/BR Voro++[contrib] WinslowUntangler Zlib
FLTK version : 1.4.0
PETSc version : 3.15.0 (real arithmtic)
OCC version : 7.6.1
MED version : 4.1.0
Packaged by : nt authority system
Web site : https://gmsh.info
Issue tracker : https://gitlab.onelab.info/gmsh/gmsh/issues
-------------------------------------------------------Gmsh launches (although not as a separate process) with the generated mesh:
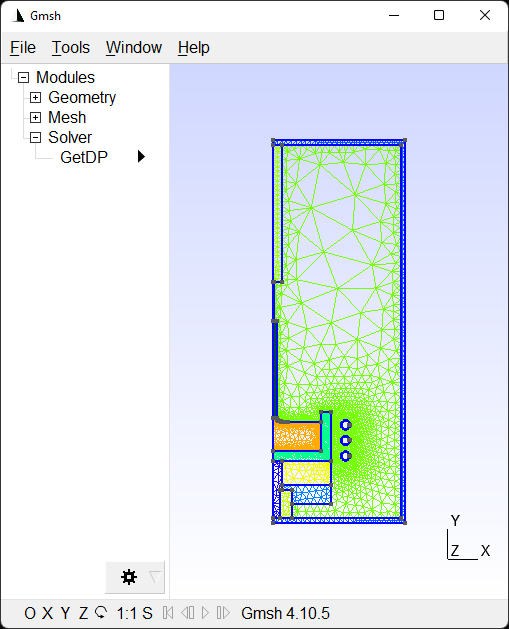
setup.py outputs the path to the mesh after closing Gmsh.
Info : Writing './simdata/_test/case.msh'...
Info : Done writing './simdata/_test/case.msh'
Gmsh model created with objectgmsh.
Shapes: ['crucible', 'melt', 'crystal', 'inductor', 'seed', 'insulation', 'crucible_adapter', 'axis_bt', 'vessel', 'axis_top', 'filling', 'bnd_melt', 'bnd_seed', 'bnd_crystal_side', 'bnd_crystal_top', 'bnd_axtop_side', 'bnd_axtop_bt', 'bnd_crucible_bt', 'bnd_crucible_outside', 'bnd_ins', 'bnd_adp', 'bnd_axbt', 'bnd_vessel_inside', 'bnd_vessel_outside', 'bnd_inductor_outside', 'bnd_inductor_inside', 'bnd_symmetry_axis', 'if_crucible_melt', 'if_melt_crystal', 'if_crystal_seed', 'if_seed_axtop', 'if_axtop_vessel', 'if_crucible_ins', 'if_ins_adp', 'if_adp_axbt', 'if_axbt_vessel']The next step will be to figure out how to simulate crystal growth using pyelmer.
Leave a Reply