Crystal Growth Simulation on Linux
Crystal Growth Simulation – Part 1 walked through setting up a mesh for running crystal simulation in Elmer on Windows. Crystal Growth Simulation Failure described the process of trying to run the actual simulation on Windows and the resulting segmentation fault. What happens if we try this same experiment on Linux? First install Elmer using the instructions on the Elmer FEM blog.
sudo apt-add-repository ppa:elmer-csc-ubuntu/elmer-csc-ppa
sudo apt-get update
sudo apt-get install elmerfem-cscI’m not sure if it’s because I’ve built other repos on my Ubuntu VM, but python3 is already installed. I still need to install pip though.
sudo apt-get install python3
sudo apt install python3-pip
pip install pyelmer
pip install objectgmshWe need to clone 2 repos to execute the crystal growth experiment. The opencgs repo is a dependency of the test-cz-induction repo.
cd ~/repos/fem/elmer/research
git clone https://github.com/nemocrys/opencgs
git clone https://github.com/nemocrys/test-cz-induction
Next, install the opencgs module using pip then set up the crystal growth simulation mesh.
cd opencgs
pip install -e .
This installation output contains an error about an incompatible numpy version but ends with a message about all the components being successfully installed.
Requirement already satisfied: kiwisolver>=1.0.1 in /home/saint/.local/lib/python3.8/site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (1.4.4)
Requirement already satisfied: fonttools>=4.22.0 in /home/saint/.local/lib/python3.8/site-packages (from matplotlib->pyelmer->opencgs==0.3.1) (4.34.4)
ERROR: pandas 1.4.3 has requirement numpy>=1.18.5; platform_machine != "aarch64" and platform_machine != "arm64" and python_version < "3.10", but you'll have numpy 1.17.4 which is incompatible.
...
Successfully installed commonmark-0.9.1 meshio-5.3.4 opencgs pandas-1.4.3 pygments-2.12.0 python-dateutil-2.8.2 pytz-2022.1 rich-12.5.1 typing-extensions-4.3.0I ignore the error and forge ahead with running the crystal simulation setup:
cd ../test-cz-induction
python3 setup.pyThis fails with an error related to numpy.
python3 setup.py
Traceback (most recent call last):
File "setup.py", line 19, in <module>
import opencgs.control as ctrl
File "/home/saint/repos/fem/elmer/research/opencgs/opencgs/__init__.py", line 5, in <module>
import opencgs.post
File "/home/saint/repos/fem/elmer/research/opencgs/opencgs/post.py", line 2, in <module>
import meshio
File "/home/saint/.local/lib/python3.8/site-packages/meshio/__init__.py", line 1, in <module>
from . import (
File "/home/saint/.local/lib/python3.8/site-packages/meshio/_cli/__init__.py", line 1, in <module>
from ._main import main
File "/home/saint/.local/lib/python3.8/site-packages/meshio/_cli/_main.py", line 5, in <module>
from . import _ascii, _binary, _compress, _convert, _decompress, _info
File "/home/saint/.local/lib/python3.8/site-packages/meshio/_cli/_ascii.py", line 4, in <module>
from .. import ansys, flac3d, gmsh, mdpa, ply, stl, vtk, vtu, xdmf
File "/home/saint/.local/lib/python3.8/site-packages/meshio/ansys/__init__.py", line 1, in <module>
from ._ansys import read, write
File "/home/saint/.local/lib/python3.8/site-packages/meshio/ansys/_ansys.py", line 14, in <module>
from .._helpers import register_format
File "/home/saint/.local/lib/python3.8/site-packages/meshio/_helpers.py", line 7, in <module>
from numpy.typing import ArrayLikeI guess I need to heed that error from pip. python – How can I upgrade NumPy? – Stack Overflow says to use the –upgrade flag.
$ pip install numpy --upgrade
Collecting numpy
Downloading numpy-1.23.1-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (17.1 MB)
|████████████████████████████████| 17.1 MB 3.6 MB/s
Installing collected packages: numpy
WARNING: The scripts f2py, f2py3 and f2py3.8 are installed in '/home/saint/.local/bin' which is not on PATH.
Consider adding this directory to PATH or, if you prefer to suppress this warning, use --no-warn-script-location.
Successfully installed numpy-1.23.1
That’s all that’s needed to get things to run. For example, setting up the mesh now works:
python3 setup.py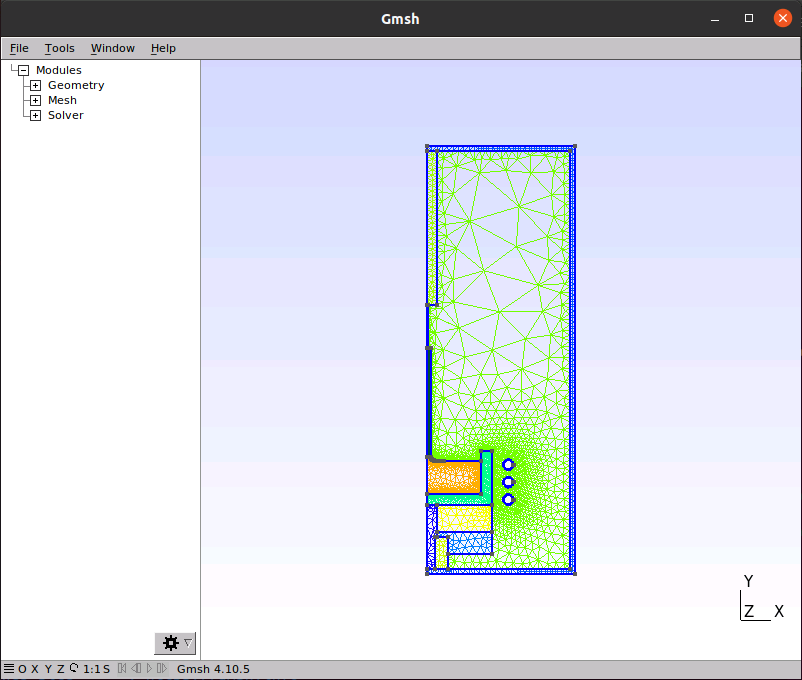
The crystal growth simulation can now be executed as well:
python3 run.pyInterestingly, the simulation completes successfully!
crucible
melt
crystal
inductor
seed
insulation
crucible_adapter
axis_bt
...
using material graphite-CZ3R6300 from self.materials_dict
using material insulation from self.materials_dict
using material steel-1.4541 from self.materials_dict
using material vacuum from self.materials_dict
Wrote sif-file.
Starting simulation ./simdata/2022-07-31_13-35_ss_test-cz-induction_vacuum ...
[] [] {'CPU-time': 55.87, 'real-time': 57.49}
Finished simulation ./simdata/2022-07-31_13-35_ss_test-cz-induction_vacuum .
Post processing...
evaluating heat fluxes
Finished post processing.This implies that the segmentation fault I ran into on Windows was specific to the Windows Elmer build.
Visualization
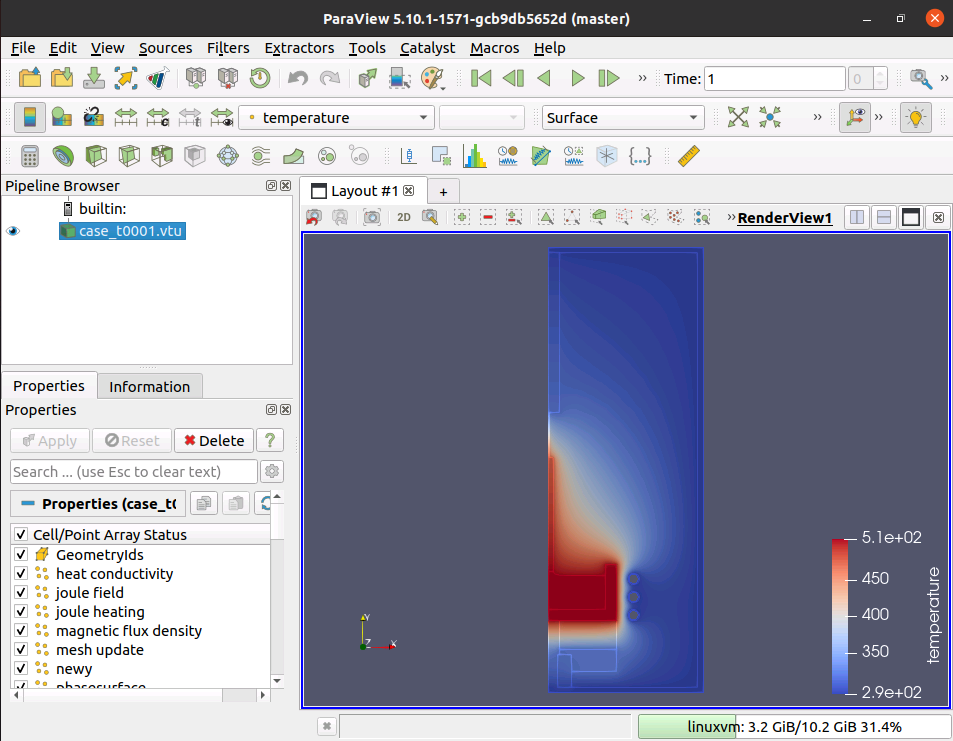
Use ParaView to visualize the results of the simulation.
sudo apt install paraviewThere are different variables that can be visualized. I selected temperature for the screenshot below. Other options included joule field, joule heating, potential im, newy, etc.
Leave a Reply